Molecular Dynamics¶
Set up environment (optional)¶
These steps are required to run this tutorial with Google Colab. To do so, uncomment and run the cell below.
This will replace pre-installed versions of numpy and torch in Colab with versions that are known to be compatible with janus-core.
It may be possible to skip the steps that uninstall and reinstall torch, which will save a considerable amount of time.
These instructions but may work for other systems too, but it is typically preferable to prepare a virtual environment separately before running this notebook if possible.
[1]:
# import locale
# locale.getpreferredencoding = lambda: "UTF-8"
# ! pip uninstall numpy -y # Uninstall pre-installed numpy
# ! pip uninstall torch torchaudio torchvision transformers -y # Uninstall pre-installed torch
# ! uv pip install torch==2.5.1 # Install pinned version of torch
# ! uv pip install janus-core[mace,visualise] data-tutorials --system # Install janus-core with MACE and WeasWidget, and data-tutorials
# get_ipython().kernel.do_shutdown(restart=True) # Restart kernel to update libraries. This may warn that your session has crashed.
To ensure you have the latest version of janus-core installed, compare the output of the following cell to the latest version available at https://pypi.org/project/janus-core/
[2]:
from janus_core import __version__
print(__version__)
0.8.6
Prepare data and modules¶
[3]:
from data_tutorials.data import get_data
get_data(
url="https://raw.githubusercontent.com/stfc/janus-core/main/docs/source/tutorials/data/",
filename=["NaCl-1040.extxyz"],
folder="../data",
)
try to download NaCl-1040.extxyz from https://raw.githubusercontent.com/stfc/janus-core/main/docs/source/tutorials/data/ and save it in ../data/NaCl-1040.extxyz
saved in ../data/NaCl-1040.extxyz
[4]:
from pathlib import Path
from ase.build import bulk
from ase.io import read, write
import numpy as np
import matplotlib.pyplot as plt
from weas_widget import WeasWidget
import yaml
from janus_core.helpers.stats import Stats
from janus_core.processing import post_process
NVT¶
In janus_core we can simulate MD in various ensembles. To start we will look at a simple NVT simulation of NaCl.
Using the ASE we can build an NaCl crystal sample. Which we can look at using Weas.
[5]:
Path("../data").mkdir(exist_ok=True)
NaCl = bulk("NaCl", "rocksalt", a=5.63, cubic=True)
NaCl = NaCl * (2, 2, 2)
write("../data/NaCl.xyz", NaCl)
v=WeasWidget()
v.from_ase(NaCl)
v
[5]:
Running the simulation¶
For our simulation we can prepare a config YAML file for the janus md command.
Here we set the ensemble and the structure.
The following 2 options specify the simulation steps to perform in MD and the MD temperature in K. The default timestep is 1 fs.
The final 2 options set the frequency to write statistics and trajectory frames.
If you have a GPU on you, then the commented out option will speed things up considerably.
[6]:
%%writefile config-nvt.yml
arch: mace_mp
ensemble: nvt
struct: ../data/NaCl.xyz
steps: 250
temp: 100
stats_every: 10
traj_every: 10
restart_every: 50
tracker: False
# If you have an Nvidia GPU.
#device: cuda
Writing config-nvt.yml
As with other janus_core functionalities the help method will show the available options along with units
[7]:
!janus md --help
Usage: janus md [OPTIONS]
Run molecular dynamics simulation, and save trajectory and statistics.
╭─ Options ────────────────────────────────────────────────────────────────────╮
│ --config TEXT Path to configuration file. │
│ --help Show this message and exit. │
╰──────────────────────────────────────────────────────────────────────────────╯
╭─ MLIP calculator ────────────────────────────────────────────────────────────╮
│ * --arch [mace|mace_mp|mace_off|m MLIP architecture to use │
│ 3gnet|chgnet|alignn|seve for calculations. │
│ nnet|nequip|dpa3|orb|mat [required] │
│ tersim|grace|esen|equifo │
│ rmer|pet_mad|uma|mace_om │
│ ol] │
│ --device [cpu|cuda|mps|xpu] Device to run calculations │
│ on. │
│ [default: cpu] │
│ --model TEXT MLIP model name, or path │
│ to model. │
│ --calc-kwargs DICT Keyword arguments to pass │
│ to selected calculator. │
│ Must be passed as a │
│ dictionary wrapped in │
│ quotes, e.g. "{'key': │
│ value}". │
╰──────────────────────────────────────────────────────────────────────────────╯
╭─ Calculation ────────────────────────────────────────────────────────────────╮
│ * --ensemble [nph|npt|nve|nv Name of │
│ t|nvt-nh|nvt-cs thermodynamic │
│ vr|npt-mtk] ensemble. │
│ [required] │
│ * --struct PATH Path of structure │
│ to simulate. │
│ [required] │
│ --steps INTEGER Number of steps │
│ in MD simulation. │
│ [default: 0] │
│ --timestep FLOAT Timestep for │
│ integrator, in │
│ fs. │
│ [default: 1.0] │
│ --temp FLOAT Temperature, in │
│ K. │
│ [default: 300.0] │
│ --equil-steps INTEGER Maximum number of │
│ steps at which to │
│ perform │
│ optimization and │
│ reset │
│ [default: 0] │
│ --minimize --no-minimize Whether to │
│ minimize │
│ structure during │
│ equilibration. │
│ [default: │
│ no-minimize] │
│ --minimize-eve… INTEGER Frequency of │
│ minimizations. │
│ Default disables │
│ minimization │
│ after beginning │
│ dynamics. │
│ [default: -1] │
│ --minimize-kwa… DICT Keyword arguments │
│ to pass to │
│ geometry │
│ optimizer, │
│ including │
│ "opt_kwargs", │
│ "filter_kwargs", │
│ and │
│ "traj_kwargs". │
│ Must be passed as │
│ a dictionary │
│ wrapped in │
│ quotes, e.g. │
│ "{'key': value}". │
│ --rescale-velo… --no-rescale-ve… Whether to │
│ rescale │
│ velocities during │
│ equilibration. │
│ [default: │
│ no-rescale-veloc… │
│ --remove-rot --no-remove-rot Whether to remove │
│ rotation during │
│ equilibration. │
│ [default: │
│ no-remove-rot] │
│ --rescale-every INTEGER Frequency to │
│ rescale │
│ velocities during │
│ equilibration. │
│ [default: 10] │
│ --post-process… DICT Keyword arguments │
│ to pass to │
│ post-processer. │
│ Must be passed as │
│ a dictionary │
│ wrapped in │
│ quotes, e.g. │
│ "{'key': value}". │
│ --correlation-… DICT Keyword arguments │
│ to pass to md for │
│ on-the-fly │
│ correlations. │
│ Must be passed as │
│ a list of │
│ dictionaries │
│ wrapped in │
│ quotes, e.g. │
│ "[{'key' : │
│ values}]". │
│ --plumed-input PATH Path to PLUMED │
│ input file. │
│ --plumed-log PATH Path for the │
│ PLUMED log file. │
│ --seed INTEGER Random seed for │
│ numpy.random and │
│ random functions. │
╰──────────────────────────────────────────────────────────────────────────────╯
╭─ Ensemble configuration ─────────────────────────────────────────────────────╮
│ --thermostat-time FLOAT Thermostat time for NPT, NPT-MTK or │
│ NVT Nosé-Hoover simulation, in fs. │
│ Default is 50 fs for NPT and NVT │
│ Nosé-Hoover, or 100 fs for NPT-MTK. │
│ --barostat-time FLOAT Barostat time for NPT, NPT-MTK or NPH │
│ simulation, in fs. Default is 75 fs │
│ for NPT and NPH, or 1000 fs for │
│ NPT-MTK. │
│ --bulk-modulus FLOAT Bulk modulus for NPT or NPH │
│ simulation, in GPa. │
│ [default: 2.0] │
│ --pressure FLOAT Pressure for NPT or NPH simulation, in │
│ GPa. │
│ [default: 0.0] │
│ --friction FLOAT Friction coefficient for NVT │
│ simulation, in fs^-1. │
│ [default: 0.005] │
│ --taut FLOAT Temperature coupling time constant for │
│ NVT CSVR simulation, in fs. │
│ [default: 100.0] │
│ --thermostat-chain INTEGER Number of variables in thermostat │
│ chain for NPT MTK simulation. │
│ [default: 3] │
│ --barostat-chain INTEGER Number of variables in barostat chain │
│ for NPT MTK simulation. │
│ [default: 3] │
│ --thermostat-substeps INTEGER Number of sub-steps in thermostat │
│ integration for NPT MTK simulation. │
│ [default: 1] │
│ --barostat-substeps INTEGER Number of sub-steps in barostat │
│ integration for NPT MTK simulation. │
│ [default: 1] │
│ --ensemble-kwargs DICT Keyword arguments to pass to ensemble │
│ initialization. Must be passed as a │
│ dictionary wrapped in quotes, e.g. │
│ "{'key': value}". │
╰──────────────────────────────────────────────────────────────────────────────╯
╭─ Heating/cooling ramp ───────────────────────────────────────────────────────╮
│ --temp-start FLOAT Temperature to start heating, in K. │
│ --temp-end FLOAT Maximum temperature for heating, in K. │
│ --temp-step FLOAT Size of temperature steps when heating, in K. │
│ --temp-time FLOAT Time between heating steps, in fs. │
╰──────────────────────────────────────────────────────────────────────────────╯
╭─ Restart settings ───────────────────────────────────────────────────────────╮
│ --restart --no-restart Whether restarting │
│ dynamics. │
│ [default: no-restart] │
│ --restart-auto --no-restart-auto Whether to infer │
│ restart file if │
│ restarting dynamics. │
│ [default: │
│ restart-auto] │
│ --restart-stem PATH Stem for restart file │
│ name. Default inferred │
│ from `file_prefix`. │
│ --restart-every INTEGER Frequency of steps to │
│ save restart info. │
│ [default: 1000] │
│ --rotate-restart --no-rotate-restart Whether to rotate │
│ restart files. │
│ [default: │
│ no-rotate-restart] │
│ --restarts-to-keep INTEGER Restart files to keep │
│ if rotating. │
│ [default: 4] │
╰──────────────────────────────────────────────────────────────────────────────╯
╭─ Output files ───────────────────────────────────────────────────────────────╮
│ --final-file PATH File to save final │
│ configuration at each │
│ temperature of similation. │
│ Default inferred from │
│ `file_prefix`. │
│ --stats-file PATH File to save thermodynamical │
│ statistics. Default inferred │
│ from `file_prefix`. │
│ --stats-every INTEGER Frequency to output │
│ statistics. │
│ [default: 100] │
│ --traj-file PATH File to save trajectory. │
│ Default inferred from │
│ `file_prefix`. │
│ --traj-append --no-traj-append Whether to append trajectory. │
│ [default: no-traj-append] │
│ --traj-start INTEGER Step to start saving │
│ trajectory. │
│ [default: 0] │
│ --traj-every INTEGER Frequency of steps to save │
│ trajectory. │
│ [default: 100] │
╰──────────────────────────────────────────────────────────────────────────────╯
╭─ Structure I/O ──────────────────────────────────────────────────────────────╮
│ --file-prefix PATH Prefix for output files, including directories. │
│ Default directory is ./janus_results, and │
│ default filename prefix is inferred from the │
│ input stucture filename. │
│ --read-kwargs DICT Keyword arguments to pass to ase.io.read. Must │
│ be passed as a dictionary wrapped in quotes, │
│ e.g. "{'key': value}". By default, │
│ read_kwargs['index'] = -1, so only the last │
│ structure is read. │
│ --write-kwargs DICT Keyword arguments to pass to ase.io.write when │
│ saving any structures. Must be passed as a │
│ dictionary wrapped in quotes, e.g. "{'key': │
│ value}". │
╰──────────────────────────────────────────────────────────────────────────────╯
╭─ Logging/summary ────────────────────────────────────────────────────────────╮
│ --log PATH Path to save logs to. │
│ Default is inferred │
│ from `file_prefix` │
│ --tracker --no-tracker Whether to save │
│ carbon emissions of │
│ calculation │
│ [default: tracker] │
│ --summary PATH Path to save summary │
│ of inputs, start/end │
│ time, and carbon │
│ emissions. Default is │
│ inferred from │
│ `file_prefix`. │
│ --progress-bar --no-progress-bar Whether to show │
│ progress bar. │
│ [default: │
│ progress-bar] │
│ --update-progress-ev… INTEGER Number of timesteps │
│ between progress bar │
│ updates. Default is │
│ steps / 100, rounded │
│ up. │
╰──────────────────────────────────────────────────────────────────────────────╯
We can begin our simulation by calling the following shell command. Which may take a few minutes on a CPU.
[8]:
! janus md --config config-nvt.yml
/home/runner/work/janus-core/janus-core/.venv/lib/python3.12/site-packages/e3nn/o3/_wigner.py:10: UserWarning: Environment variable TORCH_FORCE_NO_WEIGHTS_ONLY_LOAD detected, since the`weights_only` argument was not explicitly passed to `torch.load`, forcing weights_only=False.
_Jd, _W3j_flat, _W3j_indices = torch.load(os.path.join(os.path.dirname(__file__), 'constants.pt'))
cuequivariance or cuequivariance_torch is not available. Cuequivariance acceleration will be disabled.
Using Materials Project MACE for MACECalculator with /home/runner/.cache/mace/20231210mace128L0_energy_epoch249model
Using float64 for MACECalculator, which is slower but more accurate. Recommended for geometry optimization.
/home/runner/work/janus-core/janus-core/.venv/lib/python3.12/site-packages/mace/calculators/mace.py:197: UserWarning: Environment variable TORCH_FORCE_NO_WEIGHTS_ONLY_LOAD detected, since the`weights_only` argument was not explicitly passed to `torch.load`, forcing weights_only=False.
torch.load(f=model_path, map_location=device)
Using head Default out of ['Default']
Simulating at 100.0 K... ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 250/250 0:00:00
The outputs¶
While we are waiting for the simulation we can familiarise ourselves with the output files you will see.
The directory janus_results will be populated with the following key files named NaCl-nvt-T100.0-
stats.dattraj.extxyzfinal.extxyzmd-log.ymlmd-summary.yml
The stats file contains core statistical data on the simulation, including the step, wall-time, temperature, volume, and pressure tensor.
You could view this now while the simulation is running by the shell command
$ head janus_results/NaCl-nvt-T100.0-stats.dat
Giving something like the following
# Step | Real_Time [s] | Time [fs] | Epot/N [eV] ...
0 1.044 0.00 -3.37876019e+00 ...
10 8.772 10.00 -3.37789444e+00 ...
The first line in the stats files also includes the units used. Temperatures are all in K, lengths in Angstroms, and Pressures in GPa.
The traj and final files including the MD trajectory and the final configuration in EXTXYZ format.
md-log and md-summary give information about the running of the simulation and a summary of the options used in janus-md respectively. The latter in particular is worth a look as it also includes the default values for example the bulk_modulus.
$ cat janus_results/NaCl-nvt-T100.0-md-summary.yml
command: janus md
config:
arch: mace_mp
barostat_chain: 3
barostat_substeps: 1
barostat_time: null
bulk_modulus: 2.0
calc_kwargs: {}
...
These output files provide a full record of what you have done alongside the standard MD outputs.
Analysing the simulation¶
Once the simulation has completed we can make use of the utilities in janus_core. In particular janus_core.helpers.stats.Stats which makes viewing the stats.dat file straightforward (alternatively the file is readable by np.loadtxt).
Passing the path to our statistics we can observe the temperature, volume, and pressure resulting from the potential in NVT.
[9]:
stats = Stats("janus_results/NaCl-nvt-T100.0-stats.dat")
fig, ax = plt.subplots(ncols=3, figsize=(10,3))
ax[0].plot(stats[0], stats[5])
ax[0].set_xlabel("Step")
ax[0].set_ylabel("Temperature")
ax[1].plot(stats[0], stats[8])
ax[1].set_xlabel("Step")
ax[1].set_ylabel("Volume")
ax[2].plot(stats[0], stats[9])
ax[2].set_xlabel("Step")
ax[2].set_ylabel("Pressure")
plt.tight_layout()
Since we have been saving the trajectory every 10 steps we can also see a nice movie of our computers labour.
[10]:
traj = read("janus_results/NaCl-nvt-T100.0-traj.extxyz", index=":")
v=WeasWidget()
v.from_ase(traj)
v.avr.model_style = 1
v.avr.show_hydrogen_bonds = True
v
[10]:
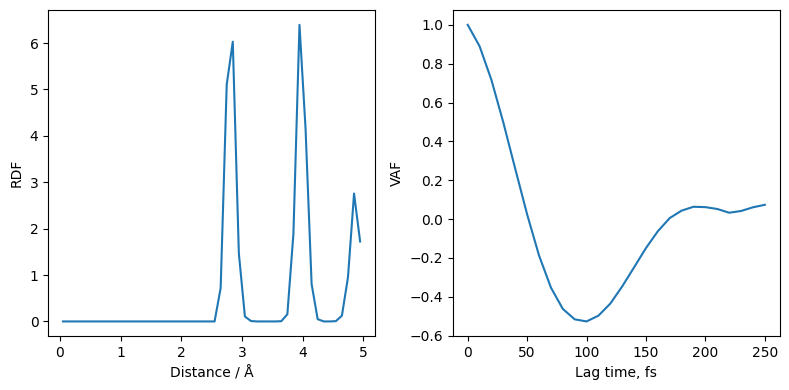
Another helper provided in janus_core is the post processing module from janus_core.processing.post_process. This enables velocity auto-correlation function and radial distribution function computation from trajectory files.
Using our data we can take a look at the RDF, and VAF
[11]:
rdf = post_process.compute_rdf(traj, rmax=5.0, nbins=50)
vaf = post_process.compute_vaf(traj)
fig, ax = plt.subplots(ncols=2, figsize=(8,4))
ax[0].plot(rdf[0], rdf[1])
ax[0].set_ylabel("RDF")
ax[0].set_xlabel("Distance / Å")
ax[1].plot(vaf[0]*10, vaf[1][0]/vaf[1][0][0])
ax[1].set_ylabel("VAF")
ax[1].set_xlabel("Lag time, fs")
plt.tight_layout()
Continuing the simulation¶
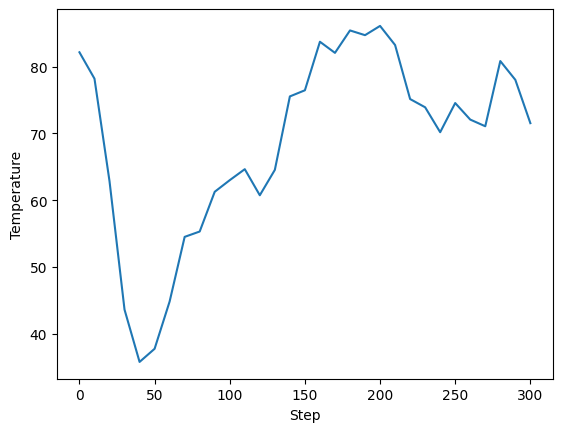
Using the final configuration we can continue the simulation easily. We can make use of our old config but override some of the values manually.
Here we indicate we want to restart with --restart, and pass our parameters from before via --config. janus_core will find our old files and begin dynamics again from the last restart file. For us this happens to be at step 250. Because we override the steps with --steps 300 we get an additional 50 simulation steps appended to our statistics and trajectory files.
[12]:
! janus md --restart --steps 300 --config config-nvt.yml
/home/runner/work/janus-core/janus-core/.venv/lib/python3.12/site-packages/e3nn/o3/_wigner.py:10: UserWarning: Environment variable TORCH_FORCE_NO_WEIGHTS_ONLY_LOAD detected, since the`weights_only` argument was not explicitly passed to `torch.load`, forcing weights_only=False.
_Jd, _W3j_flat, _W3j_indices = torch.load(os.path.join(os.path.dirname(__file__), 'constants.pt'))
cuequivariance or cuequivariance_torch is not available. Cuequivariance acceleration will be disabled.
Using Materials Project MACE for MACECalculator with /home/runner/.cache/mace/20231210mace128L0_energy_epoch249model
Using float64 for MACECalculator, which is slower but more accurate. Recommended for geometry optimization.
/home/runner/work/janus-core/janus-core/.venv/lib/python3.12/site-packages/mace/calculators/mace.py:197: UserWarning: Environment variable TORCH_FORCE_NO_WEIGHTS_ONLY_LOAD detected, since the`weights_only` argument was not explicitly passed to `torch.load`, forcing weights_only=False.
torch.load(f=model_path, map_location=device)
Using head Default out of ['Default']
Using Materials Project MACE for MACECalculator with /home/runner/.cache/mace/20231210mace128L0_energy_epoch249model
Using float64 for MACECalculator, which is slower but more accurate. Recommended for geometry optimization.
Using head Default out of ['Default']
Simulating at 100.0 K... ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━╸ 298/300 0:00:01
[13]:
stats = Stats("janus_results/NaCl-nvt-T100.0-stats.dat")
fig, ax = plt.subplots()
ax.plot(stats[0], stats[5])
ax.set_xlabel("Step")
ax.set_ylabel("Temperature")
[13]:
Text(0, 0.5, 'Temperature')
Correlations at runtime¶
Alongside post-processing of trajectory files janus_core includes a correlations module to calculate correlation functions at runtime that do not require trajectory storage.
For example beginning again from our initial structure we can compute the VAF online with the addition of correlation_kwargs.
In these options we specify a list of correlations \(\langle a b \rangle\) to compute. First the VAF by requesting a correlation with Velocity as the quantity \(a\) (\(b\) is assumed to be the same if not set) up to \(250\) correlation points (lag times). Additionally we also create a stress auto-correlation function by forming a second correlation with Stress. In this we also set the stress components to be correlated over, again \(b\) left unspecified means it is
the same as a. These extra settings will give us the correlation function \(\frac{1}{3}(\langle \sigma_{xy}\sigma_{xy}\rangle+\langle \sigma_{yz}\sigma_{yz}\rangle+\langle \sigma_{zx}\sigma_{zx}\rangle)\).
By default a correlation is updated every step. This can be controlled by the option update_frequency.
[14]:
%%writefile config-nvt-cor.yml
ensemble: nvt
struct: ../data/NaCl.xyz
arch: mace_mp
steps: 250
temp: 100
stats_every: 10
traj_every: 250
correlation_kwargs:
vaf:
a: Velocity
points: 250
saf:
a: Stress
a_kwargs: {'components': ['xy', 'yz', 'zx']}
points: 250
tracker: False
# If you have an Nvidia GPU.
#device: cuda
Writing config-nvt-cor.yml
[15]:
! janus md --config config-nvt-cor.yml
/home/runner/work/janus-core/janus-core/.venv/lib/python3.12/site-packages/e3nn/o3/_wigner.py:10: UserWarning: Environment variable TORCH_FORCE_NO_WEIGHTS_ONLY_LOAD detected, since the`weights_only` argument was not explicitly passed to `torch.load`, forcing weights_only=False.
_Jd, _W3j_flat, _W3j_indices = torch.load(os.path.join(os.path.dirname(__file__), 'constants.pt'))
cuequivariance or cuequivariance_torch is not available. Cuequivariance acceleration will be disabled.
Using Materials Project MACE for MACECalculator with /home/runner/.cache/mace/20231210mace128L0_energy_epoch249model
Using float64 for MACECalculator, which is slower but more accurate. Recommended for geometry optimization.
/home/runner/work/janus-core/janus-core/.venv/lib/python3.12/site-packages/mace/calculators/mace.py:197: UserWarning: Environment variable TORCH_FORCE_NO_WEIGHTS_ONLY_LOAD detected, since the`weights_only` argument was not explicitly passed to `torch.load`, forcing weights_only=False.
torch.load(f=model_path, map_location=device)
Using head Default out of ['Default']
Simulating at 100.0 K... ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 250/250 0:00:00
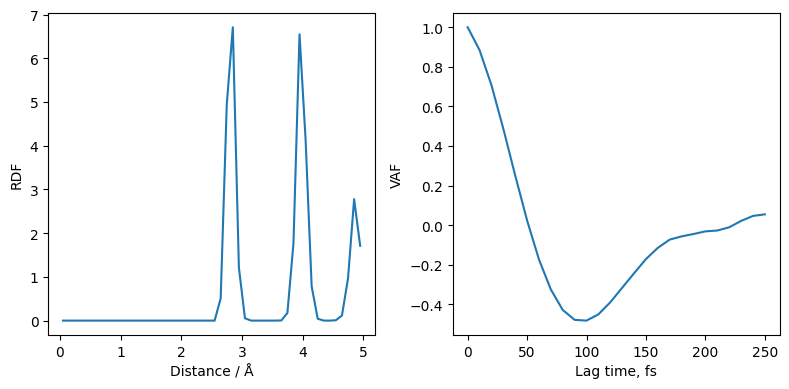
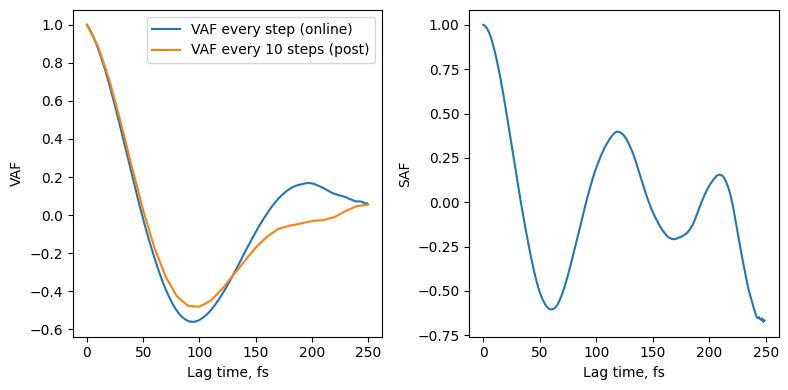
Analysing correlations¶
We can load up the new file NaCl-nvt-T100.0-cor.dat which contains the correlation function we asked for in a YAML format.
Previously we computed the VAF using the trajectory saved every 10 frames, our result here will be more accurate at the expense of a longer runtime, but we do not require any trajectory data.
[16]:
with open("janus_results/NaCl-nvt-T100.0-cor.dat", "r") as cor:
data = yaml.safe_load(cor)
vaf = data["vaf"]
saf = data["saf"]
fig, ax = plt.subplots(ncols=2, figsize=(8,4))
ax[0].plot(vaf['lags'], np.array(vaf['value'])/vaf['value'][0], label="VAF every step (online)")
ax[0].set_ylabel("VAF")
ax[0].set_xlabel("Lag time, fs")
vaf_post = post_process.compute_vaf(traj)
ax[0].plot(vaf_post[0]*10, vaf_post[1][0]/vaf_post[1][0][0], label="VAF every 10 steps (post)")
ax[0].set_ylabel("VAF")
ax[0].set_xlabel("Lag time, fs")
ax[0].legend()
ax[1].plot(saf['lags'], np.array(saf['value'])/saf['value'][0])
ax[1].set_ylabel("SAF")
ax[1].set_xlabel("Lag time, fs")
plt.tight_layout()
Heating¶
janus_core has a facility for heating or cooling during MD. This is controlled by 4 options.
temp_start: the starting temperature in Kelvin.temp_end: the final temperature in Kelvin.temp_step: the temperature increment in Kelvin.temp_time: the number of time steps at each temperature increment.
In this context the options temp and steps will apply in a normal MD run after the heating has finished.
We can make use of this to perform a naive estimation of melting in NPT by progressively increasing temperature. To keep the cell angles fixed we also apply a mask via ``ensemble_kwargs``` which are passed to ASE’s thermo-/baro-stats.
We are going to cheat by using a pre-prepared structure NaCl-1070.extxyz. This was equilibrated from ../data/NaCl.xyz in a similar way that we are about to continue but over many more steps. We can have a quick look at our starting point.
[17]:
v=WeasWidget()
structure = read("../data/NaCl-1040.extxyz")
structure.wrap()
v.from_ase(structure)
v
[17]:
We add in our heating parameters and npt options in a new config and run it.
[18]:
%%writefile config-npt-heat.yml
ensemble: npt
ensemble_kwargs:
mask: [[1,0,0],[0,1,0],[0,0,1]]
struct: ../data/NaCl-1040.extxyz
arch: mace_mp
temp_start: 1040
temp_end: 1080
temp_step: 5
temp_time: 25
stats_every: 10
traj_every: 10
tracker: False
# If you have an Nvidia GPU.
#device: cuda
Writing config-npt-heat.yml
[19]:
! janus md --config config-npt-heat.yml
/home/runner/work/janus-core/janus-core/.venv/lib/python3.12/site-packages/e3nn/o3/_wigner.py:10: UserWarning: Environment variable TORCH_FORCE_NO_WEIGHTS_ONLY_LOAD detected, since the`weights_only` argument was not explicitly passed to `torch.load`, forcing weights_only=False.
_Jd, _W3j_flat, _W3j_indices = torch.load(os.path.join(os.path.dirname(__file__), 'constants.pt'))
cuequivariance or cuequivariance_torch is not available. Cuequivariance acceleration will be disabled.
Using Materials Project MACE for MACECalculator with /home/runner/.cache/mace/20231210mace128L0_energy_epoch249model
Using float64 for MACECalculator, which is slower but more accurate. Recommended for geometry optimization.
/home/runner/work/janus-core/janus-core/.venv/lib/python3.12/site-packages/mace/calculators/mace.py:197: UserWarning: Environment variable TORCH_FORCE_NO_WEIGHTS_ONLY_LOAD detected, since the`weights_only` argument was not explicitly passed to `torch.load`, forcing weights_only=False.
torch.load(f=model_path, map_location=device)
Using head Default out of ['Default']
Simulating at 1080.0 K... ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 225/225 0:00:00
[20]:
v=WeasWidget()
structure = read("janus_results/NaCl-1040-npt-T1040.0-T1080.0-p0.0-final.extxyz")
structure.wrap()
v.from_ase(structure)
v
[20]:
Extensions¶
Viscosity¶
Simulate liquid NaCl in NVE at about 1080 K and calculate the SAF as we have done before. Use the SAF to estimate the viscosity \(\eta = \frac{V}{3k_{b}T}\int \langle \sigma_{xy}\sigma_{xy}\rangle+ \langle \sigma_{yz}\sigma_{yz}\rangle+ \langle \sigma_{zx}\sigma_{zx}\rangle dt\). Experimentally the viscosity is \(1.030\) mN s m \({}^{-2}\) [1]. Long simulations and repeats are required for accuracy, but try 1000 fs.
You will find the pre-equilibrated structure NaCl-1040.xyz useful!
[1] Janz, G.J., 1980. Molten salts data as reference standards for density, surface tension, viscosity, and electrical conductance: KNO3 and NaCl. Journal of physical and chemical reference data, 9(4), pp.791-830.
Melting¶
Try heating the NaCl crystal ../data/NaCl.xyz up to 1080 K using the temperature ramp options in NPT. To keep cell angles constant, you will want to add the options
ensemble_kwargs:
mask: [[1,0,0],[0,1,0],[0,0,1]]
temp_time: 2000 would be good if you have a GPU. You can also “cheat” by staring from NaCl-1000.xyz
Experimentally NaCl melts at about 1074 Kelvin at atmospheric pressure.